Outline¶
- Some overview of ML
- Decision trees
- Random forests and gradient boosting
- Classifiers
- Start with toy data and then some sklearn datasets
Some ML Models¶
- Linear (OLS, lasso, ridge, elastic net) -- Monday
- Trees (decision trees, random forests, gradient boosting) -- Today
- Neural networks -- in two weeks
- Support vector machines, k-nearest neighbors, etc.
Which model is best?¶
- Linear models are best if you know the data are linear.
- In general, imposing parametric assumptions is useful when you don't have much data.
- Trees are more flexible.
- Neural networks can approximate any functional relationship.
Some terminology¶
- Bagging = bootstrap aggregation
- bootstrap means to sample from the data with replacement to create random samples
- aggregation means to average the predictions of many models
- Random forests are an example of bagging
- Boosting = train a sequence of models where each model tries to correct the errors of the previous model
- Gradient boosting is an example
- Goal of bagging and boosting is to combine many weak learners to create a strong learner
- Regression in ML = prediction of a continuous variable
- Classification in ML = prediction of a categorical variable
Decision trees¶
- Random forests and gradient boosting are based on decision trees
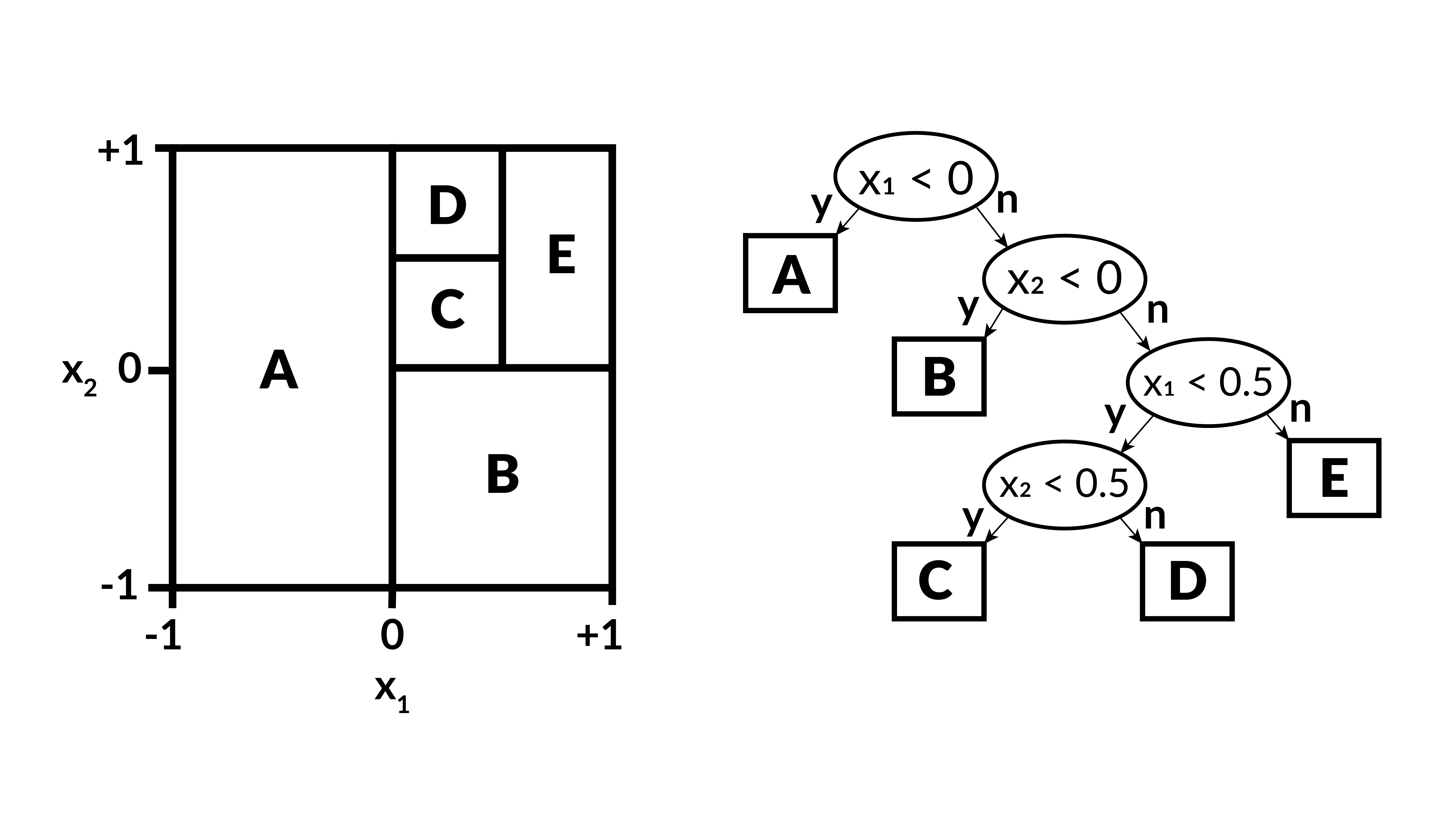
- A decision tree starts with a yes or no question.
- Depending on the answer, there is another yes or no question.
- Each question is of the form: Is variable $x_i$ greater than threshold $t_i$?
- The tree ends in a leaf node that gives the prediction.
- In regression, the tree is grown by choosing the variable and threshold that minimizes the MSE at the next step.
Example¶
Random data¶
- 10 features
- target is product of first two plus noise
- 1,500 observations (1,000 training, 200 validation, 300 test)
In [75]:
import numpy as np
np.random.seed(0) # just so we all get the same results
# full hypothetical sample
X = np.random.normal(size=(1500, 10))
y = X[:, 0] * X[:, 1] + np.random.normal(size=1500)
# randomly split into training and test samples
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=300
)
X_train0, X_val, y_train0, y_val = train_test_split(
X_train, y_train, test_size=200
)
Train a decision tree¶
- We will use a max depth of 3 for illustration.
- We will train on (X_train0, y_train0).
In [76]:
from sklearn.tree import DecisionTreeRegressor
model = DecisionTreeRegressor(max_depth=3)
model.fit(X_train0, y_train0)
Out[76]:
DecisionTreeRegressor(max_depth=3)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeRegressor(max_depth=3)
View the tree¶
- We do this only for illustration.
In [77]:
from sklearn.tree import plot_tree
import matplotlib.pyplot as plt
In [78]:
plot_tree(model, filled=True)
plt.show()
Random forest¶
- Bootstrap samples are used to create random datasets.
- A tree is fit to each random dataset.
- Average predictions of all trees to get the final prediction (for regression)
- Can choose a random subset of $n$ features at each split to select from.
- Key hyperparameters are n_estimators, max_features, and max_depth.
Gradient boosting¶
- Fit a decision tree.
- Look at its errors. Fit a new decision tree to predict the errors.
- Add a fraction (learning rate) of the error prediction to get a new prediction.
- Continue ...
- Key hyperparameters are n_estimators, learning_rate, and max_depth.
- Probably want to use xgboost library (eXtreme Gradient Boosting).
Overfitting / underfitting¶
- Decision trees can overfit if we allow them to grow too deep.
- If they are too shallow, they will underfit.
- In general,
- Model too complex or not sufficiently regularized (penalized) -> overfit
- Model too simple or too regularized -> underfit
Random forest example¶
In [79]:
from sklearn.ensemble import RandomForestRegressor
# train and validate
depths = range(2, 22, 2)
train_scores = []
val_scores = []
for depth in depths:
model = RandomForestRegressor(max_depth=depth)
model.fit(X_train0, y_train0)
train_scores.append(model.score(X_train0, y_train0))
val_scores.append(model.score(X_val, y_val))
In [80]:
# plot results
import matplotlib.pyplot as plt
plt.plot(depths, train_scores, label='train')
plt.plot(depths, val_scores, label='validation')
plt.xlabel('max_depth')
plt.ylabel('R^2')
plt.legend()
plt.show()
In [81]:
# Diebold-Mariano test
from dieboldmariano import dm_test
benchmark_predict = np.repeat(y_test.mean(), len(y_test))
best_depth = depths[np.argmax(val_scores)]
model = RandomForestRegressor(max_depth=best_depth)
model.fit(X_train, y_train) # fit using all data other than test data
model_predict = model.predict(X_test)
dm_test(y_test, model_predict, benchmark_predict, one_sided=True)
Out[81]:
(np.float64(-5.814890768275992), np.float64(7.784652577665882e-09))
Feature importances¶
- In linear models with standardized right-hand side variables, the coefficients give the importance of each variable.
- In decision trees, we can look at the feature importances, which tell us which features are used the most for splitting.
In [82]:
model.feature_importances_
Out[82]:
array([0.27893596, 0.28527914, 0.0710107 , 0.05339588, 0.04650545,
0.05839854, 0.04809928, 0.05846844, 0.05056529, 0.04934132])
Gradient boosting example with cross validation¶
In [83]:
from xgboost import XGBRegressor
from sklearn.model_selection import GridSearchCV
depths = range(2, 12, 2)
learning_rates = [0.1, 0.01, 0.001]
cv = GridSearchCV(
XGBRegressor(),
{'max_depth': depths, 'learning_rate': learning_rates}
)
# use all of our training data for cross-validation
cv.fit(X_train, y_train)
# test on test data
print(f"R2 on test data is {cv.score(X_test, y_test)}")
# see best hyperparameters
print(f"\nbest hyperparameters are {cv.best_params_}")
# feature importances
print(f"\nfeature importances are {cv.best_estimator_.feature_importances_}")
R2 on test data is 0.387006955062173
best hyperparameters are {'learning_rate': 0.1, 'max_depth': 6}
feature importances are [0.15721264 0.22603351 0.07810193 0.06738326 0.06656754 0.07817706
0.07429293 0.09016509 0.08388416 0.07818186]
Classification¶
- Binary or multi-class
- Can use lasso or ridge versions of logistic regression
- Tree-based classifiers use same syntax as regressors. Generally same hyperparameters.
- Same issues with overfitting and underfitting. Can use cross validation to choose hyperparameters as in the regression case.
- One difference is goodness of fit measure.
- Regression usually uses MSE (but can use MAE).
- Classification uses accuracy, precision, recall, F1 score, etc.
Goodness of fit in binary classification¶
- Accuracy = % correct
- Precision = % of positive predictions that are correct
- Recall = % of actual positives that are predicted correctly
- F1 score = harmonic mean of precision and recall (harmonic mean = reciprocal of average reciprocal).
- Example: cancer screening
- Precision = % of people who test positive who actually have cancer
- Recall = % of people who have cancer who test positive
- F1 score = balance between the two
Predicting probabilities¶
- Classifiers actually predict probabilities of being in each class.
- Probabilities are by default converted to class predictions as: highest probability class is the prediction.
- Can use other thresholds to convert probabilities to class predictions.
- Example: cancer screening
- If we want to catch all cases of cancer, we might set a low threshold.
- If we want to be very sure that a positive test is correct, we might set a high threshold.
Impurity¶
- In tree models, prediction is most dominant class in each leaf node.
- Choose splits to minimize impurity (Gini or entropy) rather than MSE.
- Pure means all observations in a node are of the same class.
- With $k$ classes, the Gini impurity of a group of observations is
$$ 1 - \sum_{i=1}^k p_i^2 $$
- $p_i$ is the fraction of the observations that are in class $i$.
- Perfect purity (all in 1 class) -> Gini impurity = 0
- Equal fraction in all classes -> Gini impurity = $1 - 1/k$
In [84]:
### Revised random data
import numpy as np
np.random.seed(0) # just so we all get the same results
# full hypothetical sample
X = np.random.normal(size=(1500, 10))
y = X[:, 0] * X[:, 1] + np.random.normal(size=1500)
# classify into three categories
y = np.select([y < -1, np.abs(y) <= 1, y > 1], [0, 1, 2])
# randomly split into training and test samples
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=300
)
X_train0, X_val, y_train0, y_val = train_test_split(
X_train, y_train, test_size=200
)
Decision tree classifier¶
In [85]:
from sklearn.tree import DecisionTreeClassifier
model = DecisionTreeClassifier(max_depth=3)
model.fit(X_train0, y_train0)
Out[85]:
DecisionTreeClassifier(max_depth=3)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_depth=3)
In [86]:
plot_tree(model, filled=True)
plt.show()
Random forest classifier¶
In [87]:
from sklearn.ensemble import RandomForestClassifier
depths = range(2, 22, 2)
cv = GridSearchCV(
RandomForestClassifier(),
{'max_depth': depths}
)
# use all of our training data for cross-validation
cv.fit(X_train, y_train)
# test on test data
print(f"accuracy on test data is {cv.score(X_test, y_test)}")
# see best hyperparameters
print(f"\nbest hyperparameters are {cv.best_params_}")
# feature importances
print(f"\nfeature importances are {cv.best_estimator_.feature_importances_}")
accuracy on test data is 0.63
best hyperparameters are {'max_depth': 16}
feature importances are [0.16687997 0.17617539 0.08728818 0.07671825 0.08247482 0.08678349
0.0756431 0.08334694 0.08370289 0.08098698]
More on GridSearchCV¶
- After running it, the model is automatically fit on all of the (training) data using the best parameters.
- Then methods of models are available: .predict, .score, .feature_importances_, .best_params_, .predict_proba, etc.
In [88]:
# probabilities from the best hyperparameters
cv.predict_proba(X_test)
Out[88]:
array([[0.04252002, 0.44427695, 0.51320303],
[0.04249497, 0.87481507, 0.08268996],
[0.12707821, 0.52913918, 0.34378261],
[0.09113424, 0.72451944, 0.18434631],
[0.58857143, 0.30142857, 0.11 ],
[0.15671429, 0.34228571, 0.501 ],
[0.11639415, 0.73950915, 0.14409669],
[0.1565243 , 0.60216749, 0.24130821],
[0.59 , 0.28 , 0.13 ],
[0.17531217, 0.64575661, 0.17893122],
[0.19419812, 0.3946142 , 0.41118768],
[0.35663919, 0.44286943, 0.20049138],
[0.24278765, 0.63447316, 0.12273919],
[0.46 , 0.32591837, 0.21408163],
[0.32491067, 0.41662124, 0.25846809],
[0.06 , 0.54 , 0.4 ],
[0.23504135, 0.40430821, 0.36065045],
[0.14987966, 0.6896587 , 0.16046165],
[0.23369048, 0.54793651, 0.21837302],
[0.04459183, 0.54819218, 0.40721599],
[0.08584211, 0.8200559 , 0.09410199],
[0.41106679, 0.43013389, 0.15879932],
[0.67 , 0.27 , 0.06 ],
[0.4359985 , 0.44415012, 0.11985139],
[0.22518324, 0.5662646 , 0.20855216],
[0.77117552, 0.19808374, 0.03074074],
[0.05653356, 0.76287584, 0.18059059],
[0.07837198, 0.73489694, 0.18673108],
[0.18108113, 0.58920003, 0.22971884],
[0.27722371, 0.64568479, 0.0770915 ],
[0.03138462, 0.18523077, 0.78338462],
[0.17649088, 0.66603843, 0.15747069],
[0.62574074, 0.27185185, 0.10240741],
[0.16652259, 0.70759544, 0.12588198],
[0.26 , 0.46 , 0.28 ],
[0.23666353, 0.63170657, 0.13162991],
[0.10037392, 0.75864648, 0.1409796 ],
[0.05366244, 0.73627988, 0.21005768],
[0.10352657, 0.44275362, 0.45371981],
[0.22668646, 0.55385932, 0.21945421],
[0.12194298, 0.38451754, 0.49353947],
[0.09707524, 0.74671874, 0.15620602],
[0.10085825, 0.80798098, 0.09116077],
[0.44 , 0.51 , 0.05 ],
[0.15167652, 0.65200168, 0.1963218 ],
[0.13342779, 0.67006556, 0.19650665],
[0.10193341, 0.7434428 , 0.1546238 ],
[0.45407225, 0.43729269, 0.10863506],
[0.24975823, 0.55654834, 0.19369344],
[0.26108201, 0.53017725, 0.20874074],
[0.04 , 0.160625 , 0.799375 ],
[0.42770266, 0.42677357, 0.14552377],
[0.13411315, 0.64837449, 0.21751236],
[0.05291582, 0.6624362 , 0.28464798],
[0.06071662, 0.83093849, 0.10834489],
[0.16778573, 0.70101302, 0.13120125],
[0.12696512, 0.41664632, 0.45638857],
[0.5 , 0.34 , 0.16 ],
[0.21700091, 0.67613482, 0.10686427],
[0.08329417, 0.39775286, 0.51895296],
[0.21601679, 0.58204365, 0.20193956],
[0.12481018, 0.65031152, 0.2248783 ],
[0.42 , 0.41941176, 0.16058824],
[0.23566544, 0.50007845, 0.26425612],
[0.10709408, 0.65721578, 0.23569014],
[0.09591272, 0.6157513 , 0.28833597],
[0.10737408, 0.64680552, 0.2458204 ],
[0.2627206 , 0.55482163, 0.18245777],
[0.16796877, 0.72400094, 0.10803029],
[0.10093651, 0.778541 , 0.12052249],
[0.18498855, 0.59637955, 0.2186319 ],
[0.11 , 0.21 , 0.68 ],
[0.16361369, 0.5189293 , 0.31745702],
[0.16322845, 0.62287303, 0.21389852],
[0.02984318, 0.8182376 , 0.15191922],
[0.09954976, 0.76538344, 0.13506681],
[0.34618738, 0.48593711, 0.16787552],
[0.12678403, 0.49733174, 0.37588423],
[0.17688347, 0.40075276, 0.42236377],
[0.12275916, 0.56431639, 0.31292444],
[0.40960907, 0.44481567, 0.14557526],
[0.19 , 0.2 , 0.61 ],
[0.07063276, 0.77765201, 0.15171523],
[0.20264316, 0.56246154, 0.2348953 ],
[0.06162516, 0.77173242, 0.16664242],
[0.33677725, 0.54529915, 0.1179236 ],
[0.11592915, 0.73161128, 0.15245957],
[0.28 , 0.54941176, 0.17058824],
[0.48527273, 0.40354545, 0.11118182],
[0.29 , 0.51 , 0.2 ],
[0.17950885, 0.74236597, 0.07812518],
[0.54166667, 0.3875641 , 0.07076923],
[0.18200733, 0.53510176, 0.28289091],
[0.11076253, 0.25818083, 0.63105664],
[0.05428519, 0.67017645, 0.27553836],
[0.25020074, 0.44418228, 0.30561698],
[0.1855251 , 0.68725315, 0.12722175],
[0.5124243 , 0.40378203, 0.08379367],
[0.09935845, 0.6192249 , 0.28141665],
[0.18928157, 0.60786472, 0.20285371],
[0.13054419, 0.64056355, 0.22889226],
[0.1489106 , 0.77407779, 0.07701161],
[0.09672225, 0.72620449, 0.17707326],
[0.17310836, 0.6581665 , 0.16872514],
[0.10686907, 0.71781108, 0.17531986],
[0.09407897, 0.68748801, 0.21843302],
[0.11109774, 0.36311278, 0.52578947],
[0.59280741, 0.28445185, 0.12274074],
[0.18519656, 0.5672692 , 0.24753424],
[0.11748146, 0.74761153, 0.13490701],
[0.17453454, 0.71683695, 0.10862852],
[0.16520154, 0.63509665, 0.19970181],
[0.02488248, 0.47896368, 0.49615385],
[0.28505334, 0.66240384, 0.05254282],
[0.05290842, 0.77626593, 0.17082565],
[0.13114035, 0.74681176, 0.12204789],
[0.10498413, 0.58466667, 0.31034921],
[0.20400324, 0.74115232, 0.05484444],
[0.07002198, 0.74282801, 0.18715001],
[0.08222393, 0.68963428, 0.22814179],
[0.10732907, 0.7047738 , 0.18789714],
[0.65074074, 0.28851852, 0.06074074],
[0.09610491, 0.68470641, 0.21918868],
[0.14209302, 0.42418605, 0.43372093],
[0.13282336, 0.41279744, 0.4543792 ],
[0.35492507, 0.55768242, 0.08739251],
[0.19693201, 0.51428828, 0.28877971],
[0.10833926, 0.59819229, 0.29346845],
[0.02400833, 0.36194306, 0.61404861],
[0.17130435, 0.53549872, 0.29319693],
[0.38159801, 0.49599697, 0.12240502],
[0.32315705, 0.44002999, 0.23681296],
[0.08499243, 0.79702044, 0.11798713],
[0.15509691, 0.71426357, 0.13063952],
[0.54 , 0.34964286, 0.11035714],
[0.11882099, 0.51168501, 0.369494 ],
[0.48460384, 0.47966983, 0.03572633],
[0.2725 , 0.43084231, 0.29665769],
[0.21244298, 0.37351754, 0.41403947],
[0.32621229, 0.55539351, 0.1183942 ],
[0.0477537 , 0.38542171, 0.56682459],
[0.42268734, 0.42527132, 0.15204134],
[0.06439183, 0.66436548, 0.27124269],
[0.16447506, 0.61223338, 0.22329156],
[0.27256263, 0.49699798, 0.23043939],
[0.110331 , 0.67942032, 0.21024869],
[0.12603212, 0.46124152, 0.41272636],
[0.18621955, 0.60357769, 0.21020276],
[0.29279004, 0.54235354, 0.16485642],
[0.13513325, 0.63194903, 0.23291773],
[0.06125555, 0.83030922, 0.10843523],
[0.17522408, 0.65469614, 0.17007978],
[0.10903086, 0.67688462, 0.21408452],
[0.15 , 0.28 , 0.57 ],
[0.1830891 , 0.6539702 , 0.16294071],
[0.22040662, 0.60306493, 0.17652846],
[0.09755542, 0.74074677, 0.16169781],
[0.13598741, 0.63182379, 0.2321888 ],
[0.05072665, 0.75097565, 0.1982977 ],
[0.27251282, 0.60620513, 0.12128205],
[0.0921399 , 0.63946159, 0.26839851],
[0.0742472 , 0.58207481, 0.34367799],
[0.43 , 0.45 , 0.12 ],
[0.11 , 0.35 , 0.54 ],
[0.1327701 , 0.76062341, 0.1066065 ],
[0.20973573, 0.60412836, 0.18613591],
[0.10674896, 0.63959155, 0.25365949],
[0.07481282, 0.26047179, 0.66471538],
[0.07398441, 0.74891824, 0.17709735],
[0.70074074, 0.24851852, 0.05074074],
[0.19437976, 0.56416482, 0.24145542],
[0.49950886, 0.45782733, 0.04266382],
[0.02413201, 0.66368717, 0.31218082],
[0.16851078, 0.64287895, 0.18861027],
[0.12704791, 0.72526275, 0.14768935],
[0.22514764, 0.5220798 , 0.25277256],
[0.04874408, 0.74510288, 0.20615304],
[0.16281494, 0.72334789, 0.11383717],
[0.27532563, 0.48174962, 0.24292474],
[0.28774968, 0.58400399, 0.12824633],
[0.2095607 , 0.69510056, 0.09533874],
[0.09102744, 0.53969502, 0.36927754],
[0.39850712, 0.3816092 , 0.21988369],
[0.14 , 0.31 , 0.55 ],
[0.20333333, 0.265 , 0.53166667],
[0.22176636, 0.40052087, 0.37771276],
[0.04739938, 0.69471838, 0.25788224],
[0.14432812, 0.78294944, 0.07272244],
[0.38587146, 0.4889334 , 0.12519514],
[0.20433772, 0.69011664, 0.10554564],
[0.17187369, 0.65398746, 0.17413885],
[0.78 , 0.15 , 0.07 ],
[0.12257961, 0.74615337, 0.13126702],
[0.38039216, 0.56797683, 0.05163102],
[0.06394656, 0.805254 , 0.13079944],
[0.13032787, 0.6295082 , 0.24016393],
[0.07796469, 0.79456172, 0.12747359],
[0.17757391, 0.52131697, 0.30110912],
[0.09090909, 0.36051948, 0.54857143],
[0.08579231, 0.44410196, 0.47010573],
[0.17921836, 0.59221562, 0.22856602],
[0.11941788, 0.76388804, 0.11669408],
[0.07142857, 0.42142857, 0.50714286],
[0.09621365, 0.73774949, 0.16603686],
[0.10577264, 0.37338198, 0.52084538],
[0.04078419, 0.56667585, 0.39253995],
[0.17250068, 0.63788831, 0.18961102],
[0.22925731, 0.59037446, 0.18036823],
[0.15928298, 0.62417661, 0.21654042],
[0.54 , 0.39 , 0.07 ],
[0.21220463, 0.63713317, 0.1506622 ],
[0.12125 , 0.32 , 0.55875 ],
[0.15872768, 0.68833462, 0.15293769],
[0.19444908, 0.58484172, 0.22070921],
[0.14524723, 0.66610224, 0.18865054],
[0.0808799 , 0.63378205, 0.28533805],
[0.15303742, 0.71006288, 0.1368997 ],
[0.10764199, 0.69546644, 0.19689158],
[0.20442203, 0.55743509, 0.23814288],
[0.21133333, 0.32266667, 0.466 ],
[0.26218328, 0.54632158, 0.19149515],
[0.0836852 , 0.79111902, 0.12519578],
[0.10371287, 0.75515755, 0.14112958],
[0.1733968 , 0.54388403, 0.28271917],
[0.0698116 , 0.83906176, 0.09112664],
[0.64556968, 0.28111972, 0.0733106 ],
[0.10260017, 0.73768486, 0.15971497],
[0.17345411, 0.64794441, 0.17860148],
[0.15959288, 0.6562733 , 0.18413382],
[0.13276025, 0.52148614, 0.34575361],
[0.13614711, 0.65341902, 0.21043387],
[0.35913963, 0.43601974, 0.20484062],
[0.13643673, 0.64038797, 0.2231753 ],
[0.42471111, 0.43305916, 0.14222973],
[0.48 , 0.38 , 0.14 ],
[0.07611427, 0.59997066, 0.32391507],
[0.47833333, 0.45166667, 0.07 ],
[0.23754291, 0.59714427, 0.16531283],
[0.21539211, 0.57470954, 0.20989836],
[0.43089286, 0.44910714, 0.12 ],
[0.09240802, 0.74670356, 0.16088842],
[0.07010207, 0.6308411 , 0.29905683],
[0.06226648, 0.75673072, 0.18100279],
[0.12 , 0.35333333, 0.52666667],
[0.05922991, 0.72416191, 0.21660818],
[0.05875196, 0.73656917, 0.20467887],
[0.20411747, 0.58526744, 0.2106151 ],
[0.13625391, 0.64926629, 0.21447981],
[0.12108183, 0.61207205, 0.26684612],
[0.12611048, 0.72695349, 0.14693603],
[0.04710489, 0.72019539, 0.23269971],
[0.02844424, 0.77732902, 0.19422674],
[0.25932576, 0.58556752, 0.15510673],
[0.29858192, 0.60661225, 0.09480583],
[0.25163153, 0.56565419, 0.18271429],
[0.22385537, 0.69636459, 0.07978005],
[0.13051372, 0.61901075, 0.25047553],
[0.04731831, 0.85174348, 0.10093821],
[0.11696487, 0.58787939, 0.29515573],
[0.13132205, 0.72613613, 0.14254182],
[0.18567271, 0.61034125, 0.20398603],
[0.15 , 0.24 , 0.61 ],
[0.1560896 , 0.4499911 , 0.3939193 ],
[0.0858865 , 0.78562158, 0.12849191],
[0.15112069, 0.40792729, 0.44095202],
[0.10500989, 0.74846883, 0.14652128],
[0.26695855, 0.48147821, 0.25156324],
[0.42519231, 0.37978205, 0.19502564],
[0.05394072, 0.47162925, 0.47443003],
[0.08053156, 0.63012234, 0.2893461 ],
[0.09091842, 0.82248876, 0.08659283],
[0.07089721, 0.82864483, 0.10045797],
[0.15086897, 0.64216931, 0.20696172],
[0.17746125, 0.58032312, 0.24221563],
[0.27142857, 0.32 , 0.40857143],
[0.65 , 0.25 , 0.1 ],
[0.07244725, 0.79166501, 0.13588774],
[0.11821899, 0.61978548, 0.26199553],
[0.1154415 , 0.75818729, 0.12637121],
[0.10591022, 0.77675326, 0.11733652],
[0.18776465, 0.57215845, 0.2400769 ],
[0.05 , 0.29941176, 0.65058824],
[0.15440151, 0.69619631, 0.14940219],
[0.18314723, 0.5762548 , 0.24059797],
[0.15810891, 0.63986822, 0.20202287],
[0.67 , 0.21 , 0.12 ],
[0.18232803, 0.61872229, 0.19894968],
[0.06658612, 0.81542175, 0.11799213],
[0.34760417, 0.33264093, 0.3197549 ],
[0.10525038, 0.69693532, 0.1978143 ],
[0.20957849, 0.58501744, 0.20540408],
[0.07979882, 0.81948123, 0.10071995],
[0.10996218, 0.59414023, 0.29589759],
[0.10830224, 0.76576732, 0.12593043],
[0.31284541, 0.56340729, 0.1237473 ],
[0.20576617, 0.63130563, 0.1629282 ],
[0.11735473, 0.55663749, 0.32600778],
[0.15244814, 0.67779776, 0.1697541 ],
[0.08824957, 0.65032825, 0.26142218],
[0.13914556, 0.70207185, 0.15878259]])
Confusion matrix and ROC curve¶
- Confusion matrix is a table of actual vs. predicted classes. Reveals where the errors are.
- ROC curve is a plot of true positive rate vs. false positive rate for different thresholds of the predicted probabilities.
- Used for binary classification. Can extend to multiclass by creating dummies.
- AUC is the Area Under the (ROC) Curve. AUC = 1 is perfect, AUC = 0.5 is random.
- Useful for picking an optimal threshold, given a model.
In [89]:
from sklearn.metrics import ConfusionMatrixDisplay
ConfusionMatrixDisplay.from_estimator(
estimator=cv,
X=X_test,
y=y_test
)
plt.show()
Binary classification example¶
- Target = dummy for y > 0
In [90]:
### Second revision of data
import numpy as np
np.random.seed(0) # just so we all get the same results
# full hypothetical sample
X = np.random.normal(size=(1500, 10))
y = X[:, 0] * X[:, 1] + np.random.normal(size=1500)
# classify into two categories
y = 1 * (y > 0)
# randomly split into training and test samples
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=300
)
X_train0, X_val, y_train0, y_val = train_test_split(
X_train, y_train, test_size=200
)
In [91]:
# exact same code as for multi-class classification
depths = range(2, 22, 2)
cv = GridSearchCV(
RandomForestClassifier(),
{'max_depth': depths}
)
# use all of our training data for cross-validation
cv.fit(X_train, y_train)
# test on test data
print(f"accuracy on test data is {cv.score(X_test, y_test)}")
# see best hyperparameters
print(f"\nbest hyperparameters are {cv.best_params_}")
# feature importances
print(f"\nfeature importances are {cv.best_estimator_.feature_importances_}")
accuracy on test data is 0.61
best hyperparameters are {'max_depth': 10}
feature importances are [0.15459823 0.14353626 0.0949279 0.08389458 0.08679904 0.09218103
0.08275622 0.09064638 0.08656148 0.08409889]
In [92]:
from sklearn.metrics import RocCurveDisplay
RocCurveDisplay.from_estimator(
estimator=cv,
X=X_test,
y=y_test,
)
plt.show()
Saving models¶
To save a trained model so you can use it again without training again, use joblib.dump(model, 'filename').
import joblib
joblib.dump(model, 'filename')or
from joblib import dump
dump(model, 'filename')In [93]:
# saving
from joblib import dump
dump(cv, 'mymodel.joblib')
Out[93]:
['mymodel.joblib']
In [94]:
# loading and reusing
from joblib import load
cv = load('mymodel.joblib')
cv.predict(X_test)
Out[94]:
array([1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0,
0, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 0,
1, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1,
1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1,
0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1,
0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 0, 0,
1, 0, 0, 1, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1,
0, 0, 1, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0,
1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1,
1, 1, 1, 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 0,
1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 0, 0, 0, 0, 1, 0,
1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1,
1, 0, 0, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0,
1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0])
Ask Julius¶
- Get the California house price data, run random forest regressor on it, and show the feature importances.
- Get the digits data, show the images, run a classifier on it, and show the confusion matrix.
- Get the breast cancer data, run a classifier on it, and show the confusion matrix and ROC curve.